|
Final year Dual Degree (B.Tech + M.Tech) student in Electrical Engineering at IIT Bombay, with a minor in Artificial Intelligence, Machine Learning & Data Science. I am currently working on Vision-Language Models (VLMs) at MeDAL Lab under the guidance of Prof. Amit Sethi. I have worked on cutting-edge industrial problems in ML, NLP, Generative AI, LLMs, and Computer Vision at Fujitsu Research, Intel, and Swiggy. I am also proficient in DSA and programming, and skilled in Signal Processing and Communication Systems. Skills:
I have a strong interest in Generative AI, NLP, Computer Vision, and Machine Learning and enjoy working on projects in these areas. I’m excited to apply my skills to impactful projects. Interested in full-time opportunities in Machine Learning, Data Science, and Software Engineering starting Summer 2026. Publications:
Education:
Email / Google Scholar / GitHub / LinkedIn / X |

|

|

|

|

|

|
|
|
|
|
|
Fujitsu Research May 2025 - July 2025
|
|
|
Intel Corporation July 2024 - May 2025
|
|
|
Swiggy July 2024 - August 2024
|
|
|
IBM May 2024 - June 2024
|
|
|
GMAC Intelligence December 2023 - January 2024
| |
|
MURVEN Design Solutions December 2022 - April 2023
|
|
|
|
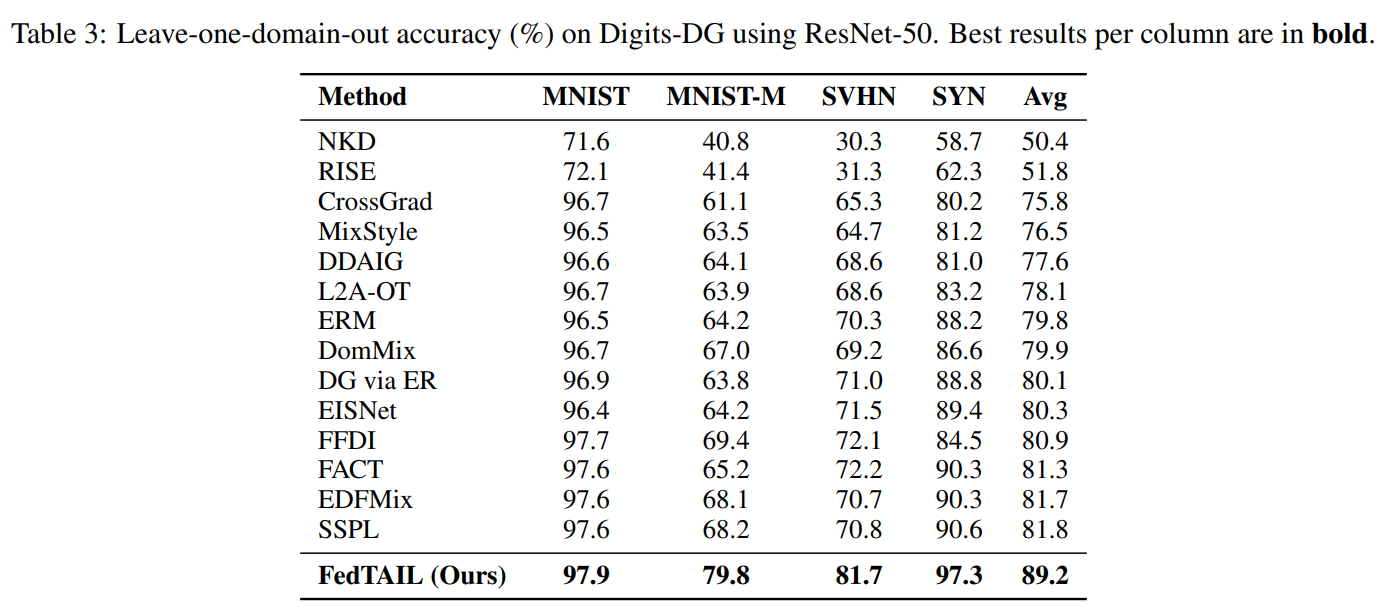
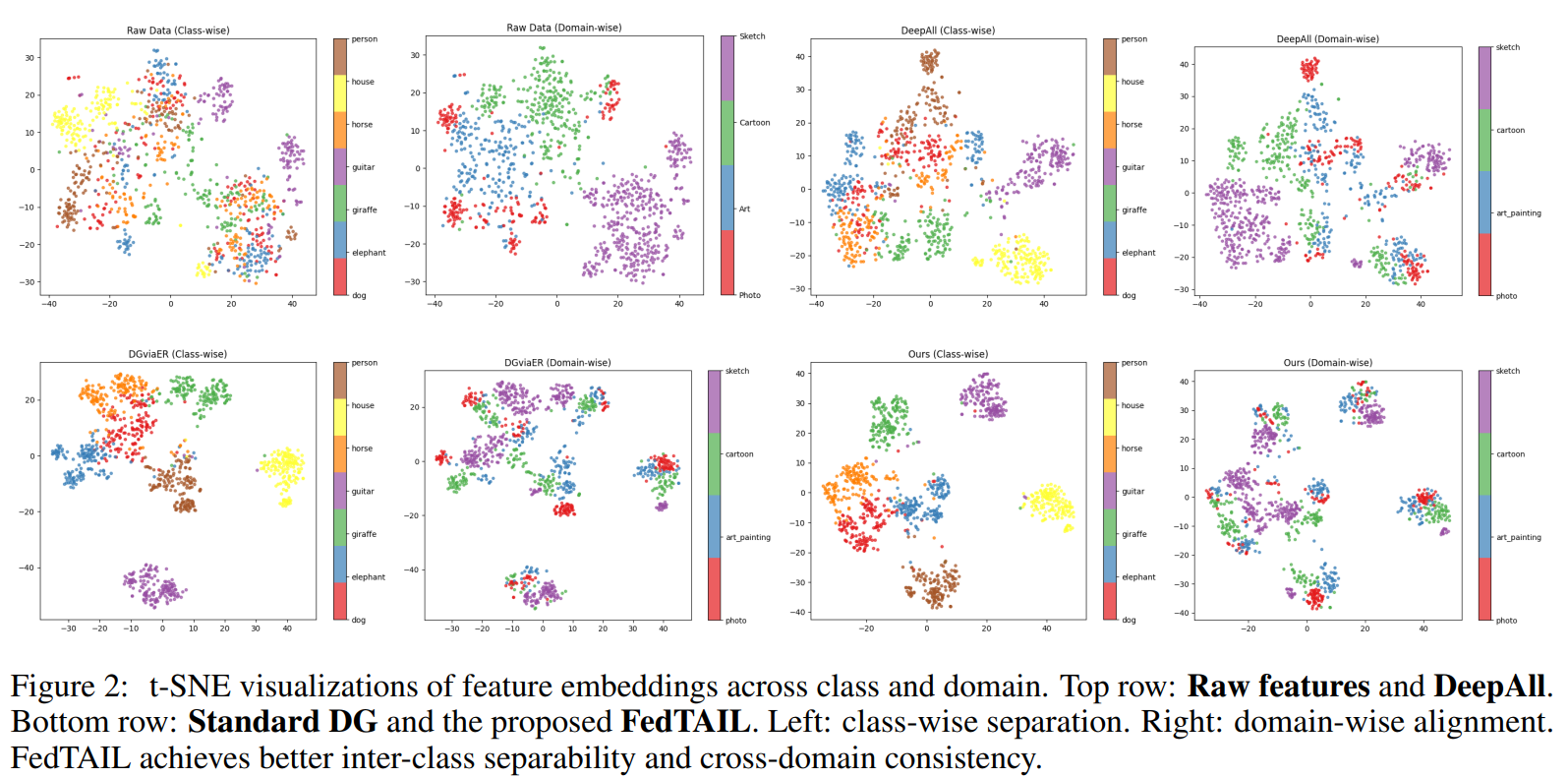
42nd International Conference on Machine Learning · ICML 2025 Vancouver, Canada We present FedTAIL, a federated domain generalization framework designed to tackle domain shifts and long-tailed class distributions. By aligning gradients across objectives and dynamically reweighting underrepresented classes using sharpness-aware optimization, our method achieves state-of-the-art performance under label imbalance. FedTAIL enables scalable and robust generalization in both centralized and federated settings. |
|
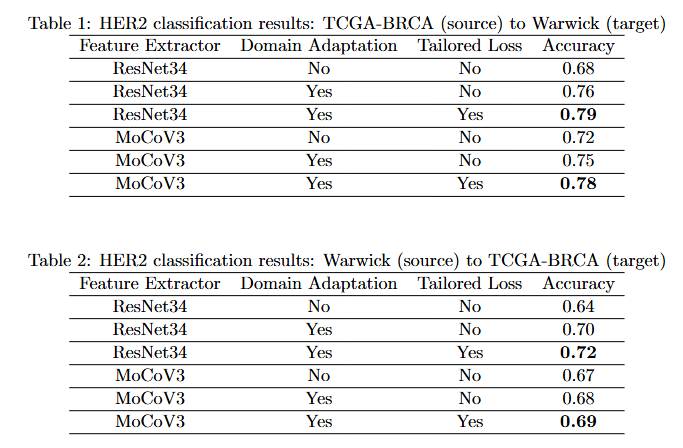
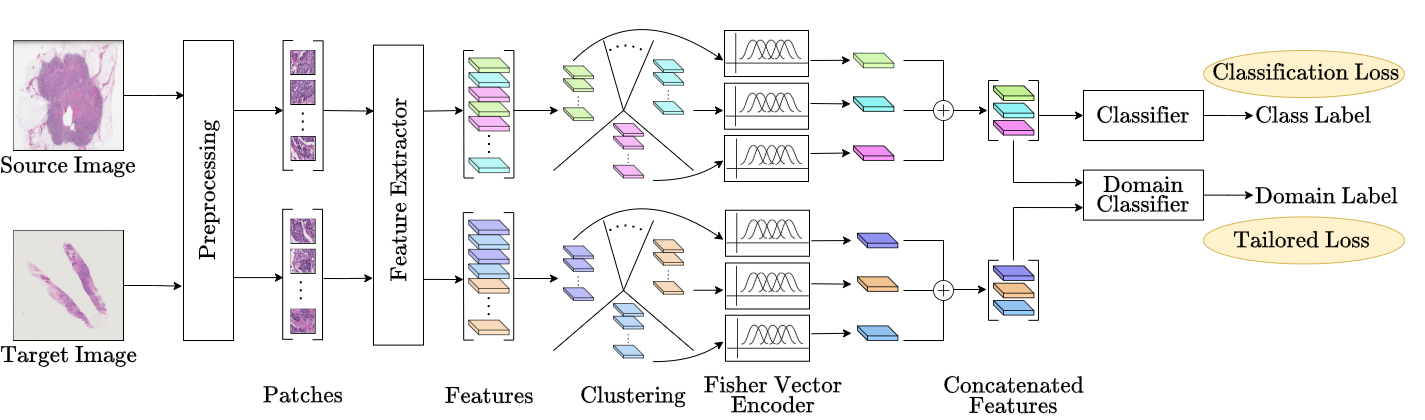
8th Medical Imaging with Deep Learning · MIDL 2025 Salt Lake City, USA We introduce a domain adaptation framework for Whole Slide Image (WSI) classification that combines self-supervised learning, clustering, and Fisher Vector encoding. By extracting MoCoV3-based patch features and aggregating them via Gaussian mixture models, our method forms robust slide-level representations. Adversarial training with a hybrid PLMMD-MCC loss enables effective domain alignment, achieving strong performance on cross-domain HER2 classification tasks, even under label noise. |
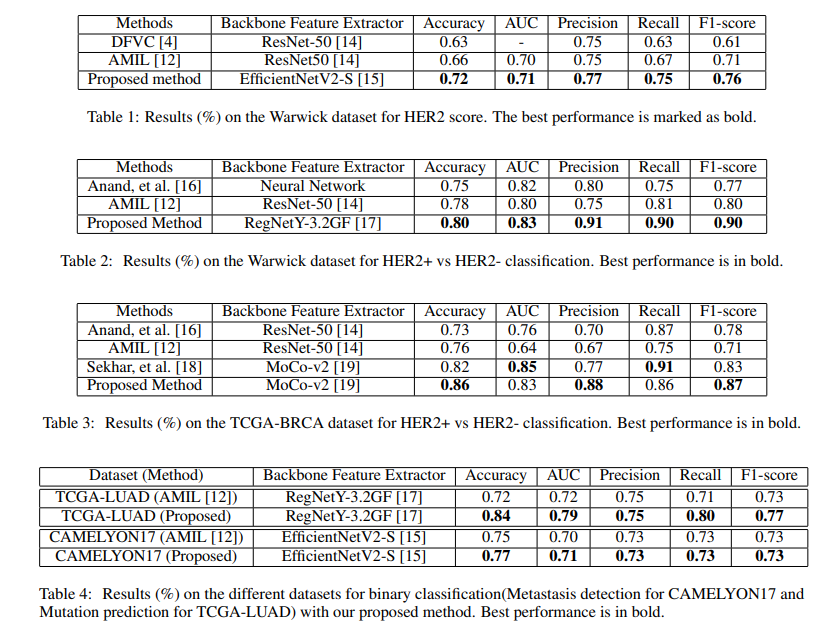
|
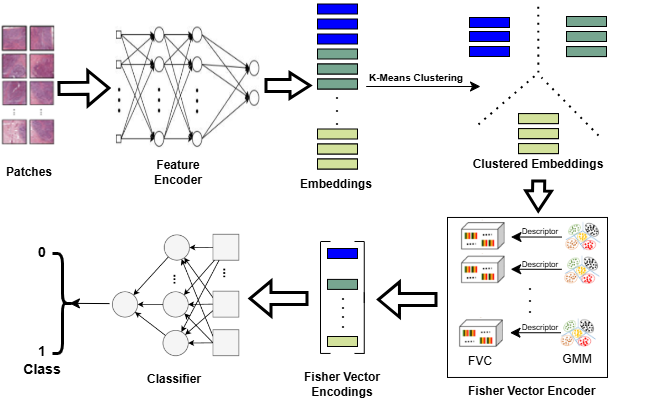
IEEE 22nd International Symposium on Biomedical Imaging · ISBI 2025 Texas, USA We propose a scalable method for whole slide image (WSI) classification that combines patch-based deep feature extraction, clustering, and Fisher Vector encoding. By modeling clustered patch embeddings with Gaussian mixture models, our approach generates compact yet expressive slide-level representations. This enables robust and accurate WSI classification while efficiently capturing both local and global tissue structures. |

|
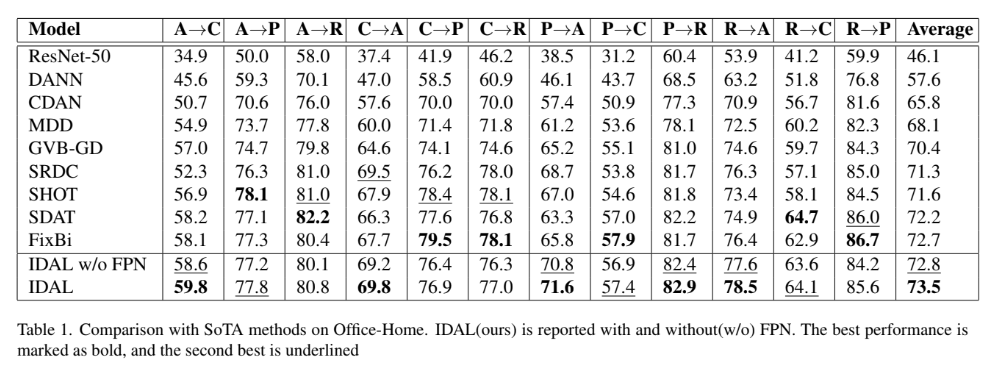
27th International Conference on Pattern Recognition · ICPR 2024 Kolkata, India We propose a novel unsupervised domain adaptation (UDA) approach for natural images that combines ResNet with a feature pyramid network to capture both content and style features. A carefully designed loss function enhances alignment across domains with multi-modal distributions, improving robustness to scale, noise, and style shifts. Our method achieves superior performance on benchmarks like Office-Home, Office-31, and VisDA-2017, while maintaining competitive results on DomainNet. |
|
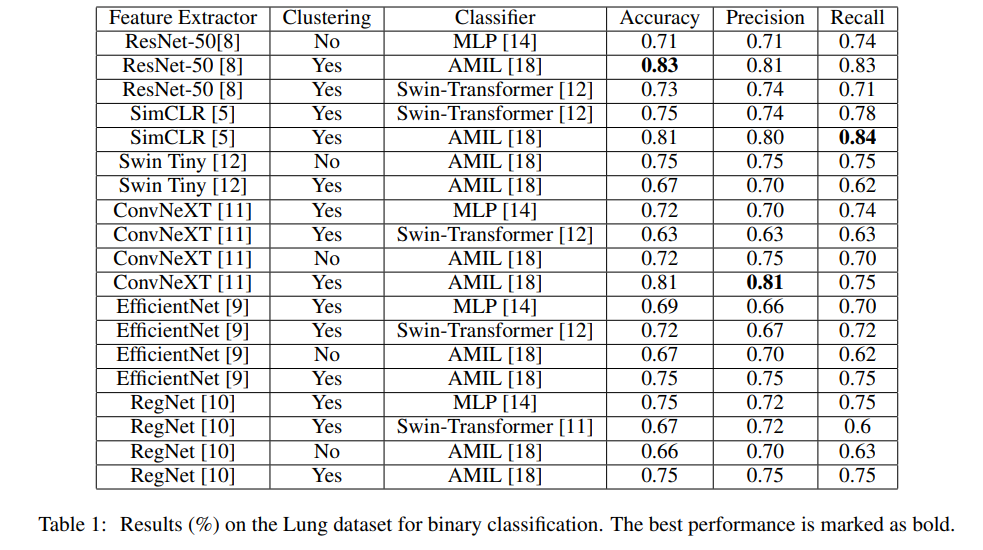
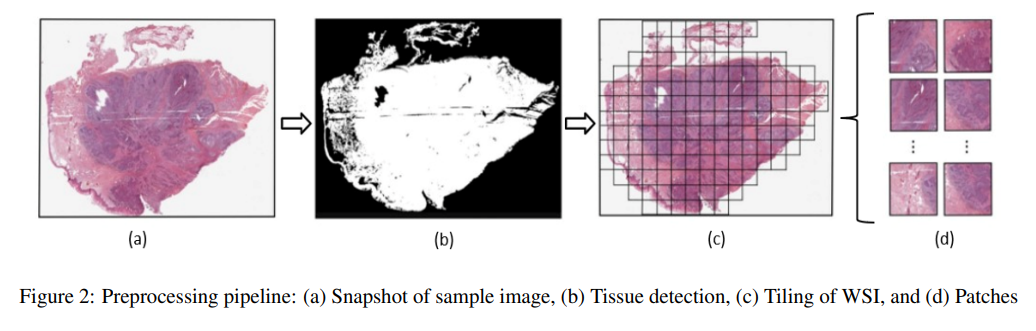
IEEE 24th International Conference on Bioinformatics and Bioengineering · BIBE 2024 Kragujevac, Serbia We propose an efficient WSI analysis framework that leverages diverse encoders and a specialized classification model to produce robust, permutation-invariant slide representations. By distilling a gigapixel WSI into a single informative vector, our method significantly improves computational efficiency without sacrificing diagnostic accuracy. This scalable approach enables effective utilization of WSIs in digital pathology and medical research. |
|
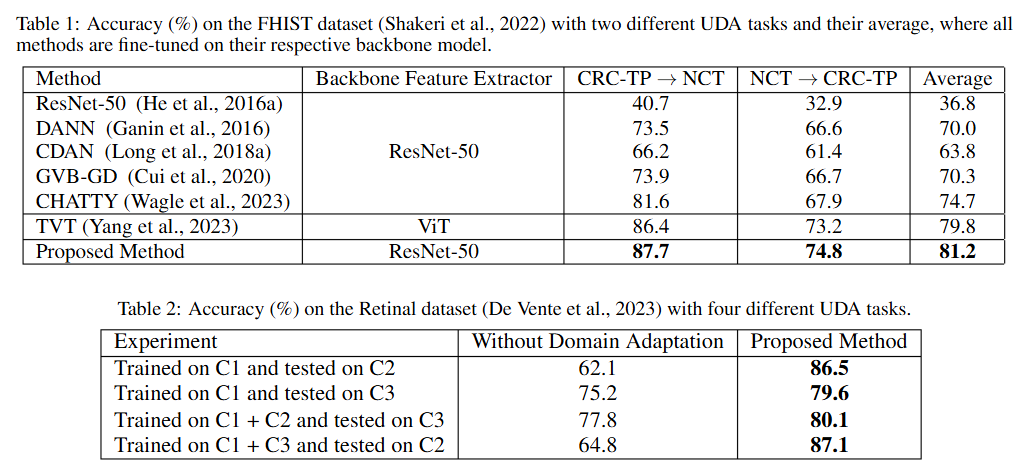
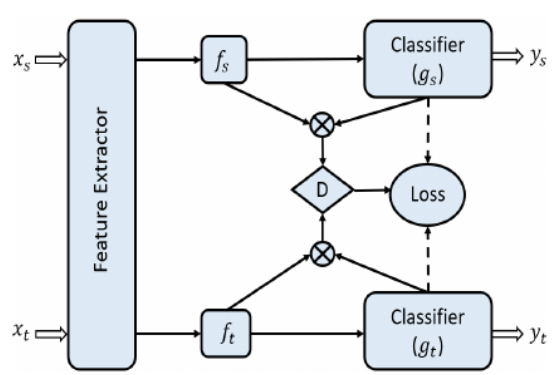
11th International Conference on BioImaging · BioImaging 2024 Rome, Italy Received the Best Student Paper Award We propose a novel approach for unsupervised domain adaptation designed for medical images like H&E-stained histology and retinal fundus scans. By leveraging texture-specific features such as tissue structure and cell morphology, DAL improves domain alignment using a custom loss function that enhances both accuracy and training efficiency. Our method outperforms ViT and CNN-based baselines on FHIST and retina datasets, demonstrating strong generalization and robustness. |
|
Website template borrowed from here. |


